Introduction to the dcsbm package
introduction.RmdYou can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("mpff/dcsbm")
library(dcsbm)1. No degree correction
We can simulate a network with a clear block structure using the
planted partition model via sample_ppm. And then estimate a
SBM using the dcsbm function.
G <- sample_ppm(60, 0.9, 5, 3, directed = TRUE, loops = TRUE)
model <- dcsbm(G, n.sweeps = 1, verbose = FALSE)
plot(G, vertex.color = model$best_partition, vertex.label = NA, sub = "Estimated partition")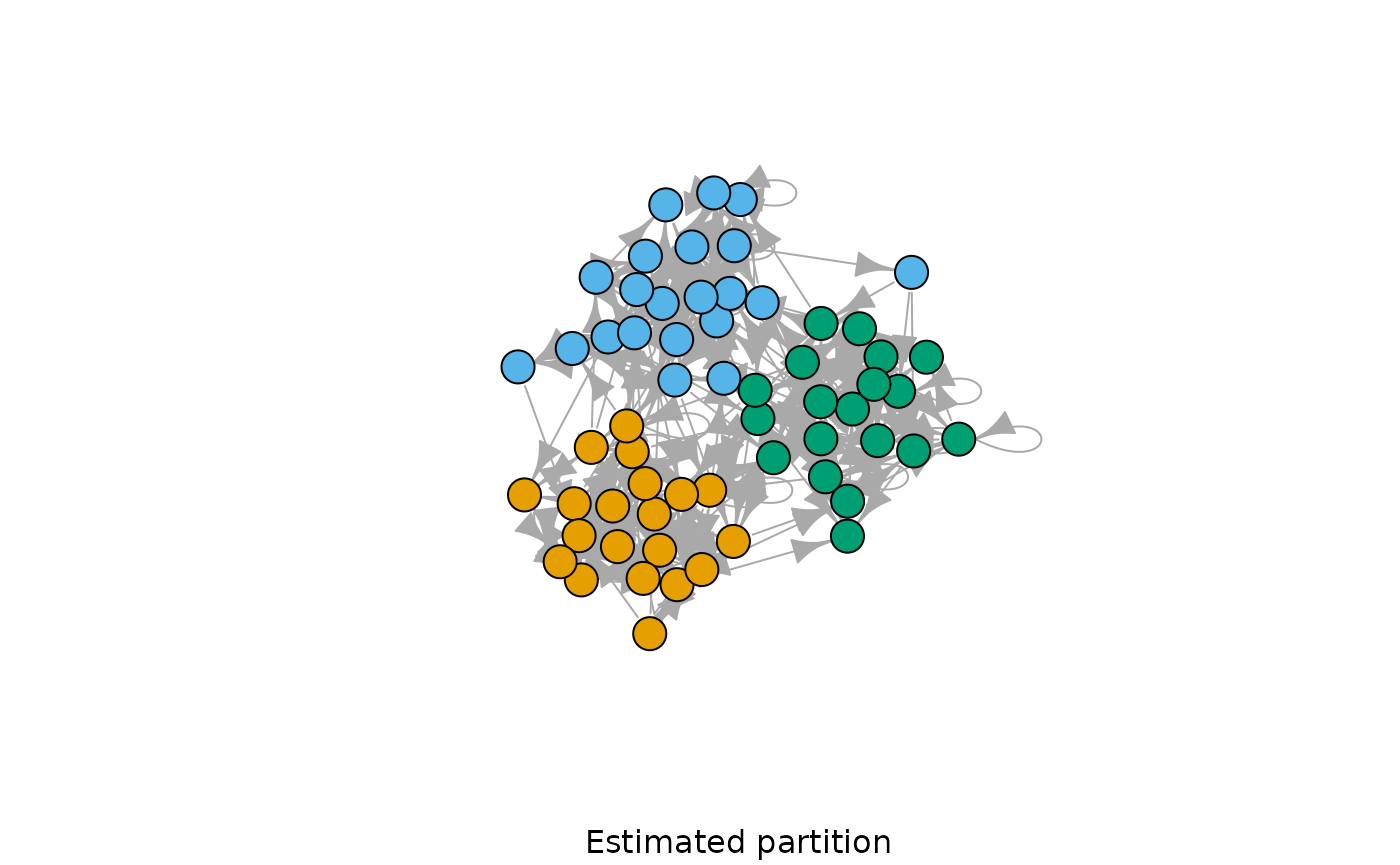
2. With degree correction
We can simulate a planted partition network with additional degree
variability inside each block via sample_dcppm. And then
estimate a degree corrected SBM by using dcsbm and setting
degree_correction = TRUE.
G <- sample_dcppm(60, 0.9, 5, 3, k_coef = 2, directed = TRUE, loops = TRUE)
model <- dcsbm(G, degree_correction = TRUE, n.sweeps = 5, verbose = FALSE)
plot(G, vertex.color = model$best_partition, vertex.label = NA, sub = "Estimated partition")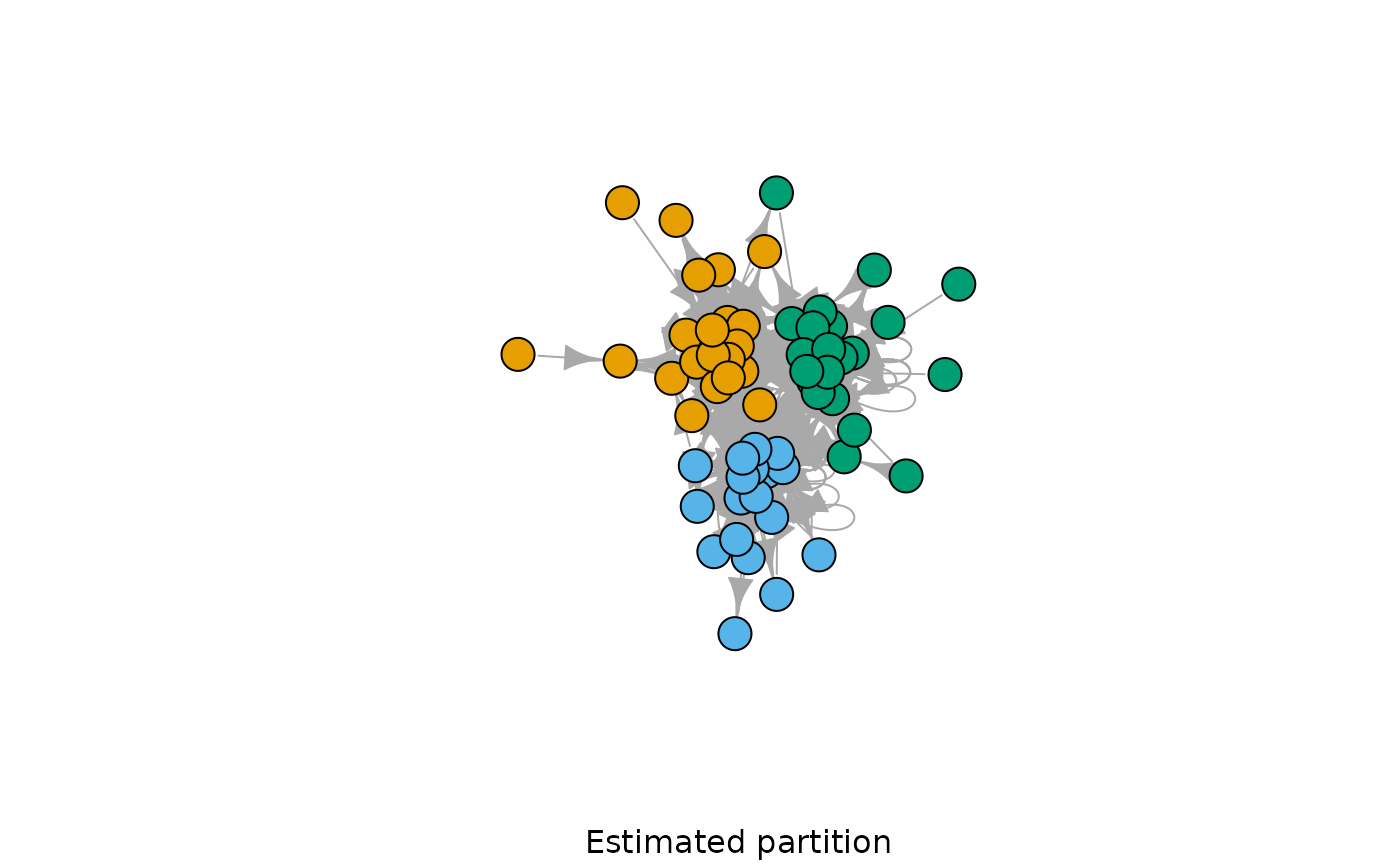
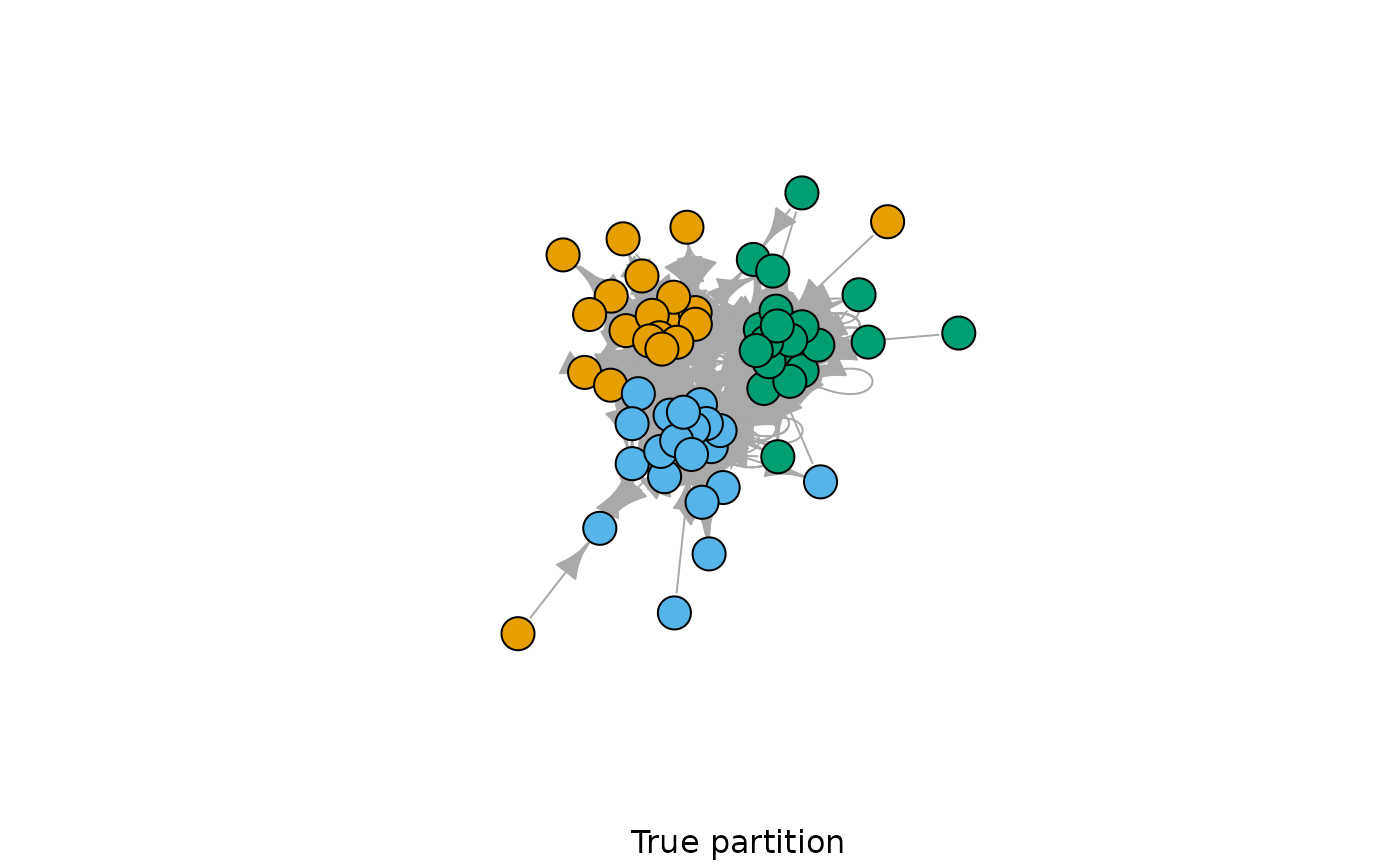
plot(G, vertex.color = c(rep(1,20), rep(2,20), rep(3,20)), vertex.label = NA, sub = "True partition")